This page contains a description of the research tasks in work package 5 "Estimating mesopelagic resources and their resilience".
Overview of tasks in work package 5
This work package will use a series of models to produce improved estimates of stock biomass, and to explore the consequences of possible future harvesting and climate change.
There are three tasks in this work package. These involve, firstly, bringing together existing data, and outputs from the field work (work package 3 and work package 4), that will be used to parameterize and test the models. Secondly, assessment models will be used to produce the stock biomass estimates. And finally spatial population models and ecosystem models will be used to explore the harvesting and climate change secanrios. Outputs from all the models will feed in to the work packages dealing with management (work package 6) and governance (work package 7).
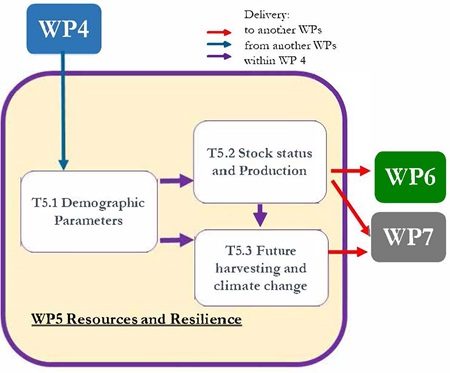
Flows of information and data to, from and within work package 5.
Task 5.1. Estimation of key demographic parameters
Lead by Technical University of Denmark
This task will collate published information, and outputs from the work packages on fishing technology development (work package 3) and the field campaign (work package 4), to provide parameters and fitting data for the models used in tasks 2 and 3 (described below).
Key data for the task 2 assessment models include population length distributions from catches and acoustic surveys; the proportion of the stock mature at size; weight-length relationships; and selectivity of fishing gear. Other required life-history parameters, such as those governing energy allocation to activity and reproduction, will be obtained from established life-history scaling rules. Age -given-length data will be compiled for vertical life-table analysis in order to provide estimates independent of those of the purely size-based methods.
The task 3 spatial and ecosystem models will require the additional quantification of growth and/or mortality rates as functions of temperature and/or food availability, requiring stomach content data and survey estimates of prey availability.
Two workshops will be organised to facilitate the interaction between the empirical and modelling work.
Task 5.2. Improved stock status and production estimates
Lead by Technical University of Denmark
The S6 model (Kokkalis et al. 2017; see bottom of page) will be used to estimate natural mortality rates, recruitment, and relative stock status (biomass over biomass at maximum sustainable yield), including uncertainty estimates, for target species (Benthosema glaciale and Maurolicus muelleri) in the northeastern Atlantic.
The model is a trait-based size spectrum, with a minimum data requirement of a stock size distribution. Normally the size distribution would be from a commercial catch, but for our target species current fishing is effectively zero, and the length distributions will be from trial catches (work package 3) and surveys (work package 4). This will permit the direct estimation of natural mortality, and relative biomass.
The trait parameters (growth rate, activity costs, investment in reproduction, and maturity ogive) will initially be based on life-history invariants and existing literature on related species, before being refined on the basis of new data from work package 4.
Task 5.3. Explore possible outcomes under future harvesting and climate change scenarios
Lead by University of Strathclyde
Three models will be used in this task.
Firstly, the StrathSPACE space and size structured population simulation scheme (Speirs et al. 2006, Heath et al. 2014, Wilson et al. 2016; see bottom of page) will be used for single species spatial modelling of the target species Benthosema glaciale and Maurolicus muelleri. The models will integrate the effects of birth, death, growth, and transport by water currents, at a resolution of ~20km over the whole North Atlantic basin, driven by flow, temperature, and zooplankton outputs from the NEMO-MEDUSA ocean biogeochemical model. StrathSPACE will be used to test the population sensitivity to spatial patterns of harvesting rate, maximum sustainable yield harvesting, balanced harvesting, and climate change (1980-2100 IPCC RCP8.5).
Secondly, food-web interactions will be explored using Individual Based Modelling (IBM) of mesopelagic fish and krill in NORWECOM.E2E (Utne et al. 2012, Skaret et al. 2014; see bottom of page) using salinity, temperature, oxygen, and velocity fields from the ROMS model. The model resolution is 10-20 km grid horizontally and 21 vertical layers, covering the Nordic Seas. This model will be used to study the mesopelagic regulation of carbon fluxes under harvesting rate and climate scenarios.
Finally, the SEAPODYM model (Dragon et al. 2015; see bottom of page) to quantify how fishing mesopelagics may impact feeding, spawning habitats, and natural mortality of commercially important top predators (Atlantic albacore). Using acoustic data (mainly from COPERNICUS CMEMS, GREENUP, and EU MESOPP projects) and survey biomass estimation (work package 4) and other case studies (e.g. Davison et al. 2015 in California CalCOFI region; see bottom of page), SEAPODYM will permit the estimation of biomass and dynamics of mesopelagic functional groups at a global scale (Lehodey et al 2010, 2015; see bottom of page). A vertical 1D coupling to a biogeochemical model (PISCES-SEAPODYM, in collaboration with Marion Gehlen, Laboratoire des Sciences du Climat et de l’Environnement) will permit a comparison with the carbon budget in PISCES (Aumont et al. 2015; see bottom of page) and an analysis of carbon fluxes due to mesopelagic functional groups under climate change.
References
Aumont O., Ethé C., Tagliabue A., Bopp L., & Gehlen M. (2015) PISCES-v2: an ocean biogeochemical model for carbon and ecosystem studies. Geoscientific Model Development 8, 2465–2513.
Davison, P., Lara-Lopez, A., Koslow, J.A. (2015) Mesopelagic fish biomass in the southern California current ecosystem. Deep-Sea Research Part II-Topical Studies in Oceanography 112, Special Issue S1, 129-142.
Dragon A.C., Senina I., Conchon A., Titaud O., Arrizabalaga H., & Lehodey P. (2015) Modeling spatial population dynamics of North Atlantic Albacore tuna under the influence of both fishing and climate variability. Canadian Journal of Fisheries and Aquatic Sciences 72, 864-878.
Heath, M.R., Culling, M.A., Crozier, W.W., Fox, C.J, Gurney, W.S.C., Hutchinson, W.F., Nielsen E.E., O’Sullivan, M., Preedy, K.F., Righton, D.A., Speirs, D.C., Taylor, M.I., Wright, P.J., & Carvalho, G.R. (2014) Combination of genetics and spatial modelling highlights the sensitivity of cod (Gadus morhua) population diversity in the North Sea to distributions of fishing. ICES Journal of Marine Science 71, 794-807.
Kokkalis, A., Eikeset, A.M., Thygesen, U.H., Steingrund, P., & Anderson, K.H. (2017) Estimating uncertainty of data limited stock assessments. ICES Journal of Marine Science 74, 69-77.
Lehodey P., Murtugudde R., Senina I. (2010). Bridging the gap from ocean models to population dynamics of large marine predators: a model of mid-trophic functional groups. Progress in Oceanography, 84: 69–84.
Lehodey, P., Conchon, A., Senina, I., Domokos, R., Calmettes, B., Jouanno, J., Hernandez, O., & Kloser, R. (2015) Optimization of a micronekton model with acoustic data. ICES Journal of Marine Science 72, 1399-1412.
Skaret, G., Dalpadado, P., Hjøllo, S. S., Skogen, M. D., & Strand, E. (2014) Calanus finmarchicus abundance, production and population dynamics in the Barents Sea in a future climate. Progress in Oceanography 125, 26-39.
Speirs, D.C., Gurney, W.S.C., Heath, M.R., Horbelt, W., Wood, S.N., & de Cuevas, B.A. (2006) Ocean-scale modelling of the distribution, abundance, and seasonal dynamics of the copepod Calanus finmarchicus. Marine Ecology Progress Series 313, 173-192.
Utne, K. R., Hjøllo, S. S., Huse, G., and Skogen, M. D. 2012. Estimating the consumption of Calanus finmarchicus by planktivorous fish in the Norwegian Sea using a fully coupled 3D model system. Marine Biology Research 8, 527-547.
Wilson, R.J., Heath, M.R., & Speirs, D.C. (2016) Spatial Modelling of Calanus finmarchicus and Calanus helgolandicus: Parameter Differences Explain Differences in Biogeography. Frontiers in Marine Science 3, Article 3, doi:10.3389/fmars.2016.00157.